REX-GECO Structure
Simulations and homology modeling to determine REX-GECO protein structure.
Introduction
REX-GECO is a fluorescent calcium biosensor protein developed in 2014. A protein that glows red in the presence of calcium, it is distinctive for how drastically its fluorescence color changes when it binds to calcium. This large color difference is beneficial for bioimaging because it reduces the color interference between the calcium bound and calcium free protein forms. Therefore, it is useful to understand what structural features contribute to this color property so that future fluorescent proteins can be designed with these features in mind. In this project, I estimated and analyzed the structure of this protein using homology modeling and molecular dynamics (MD) simulations.
Homology Modeling
The REX-GECO protein consists of three components: a fluorescent protein region, a Calmodulin (CaM) calcium binding region, and a myosin light chain kinase (M13) fragment that interacts with CaM during calcium binding. The figure below illustrates the different protein regions, and how they change conformation when calcium binds to CaM.
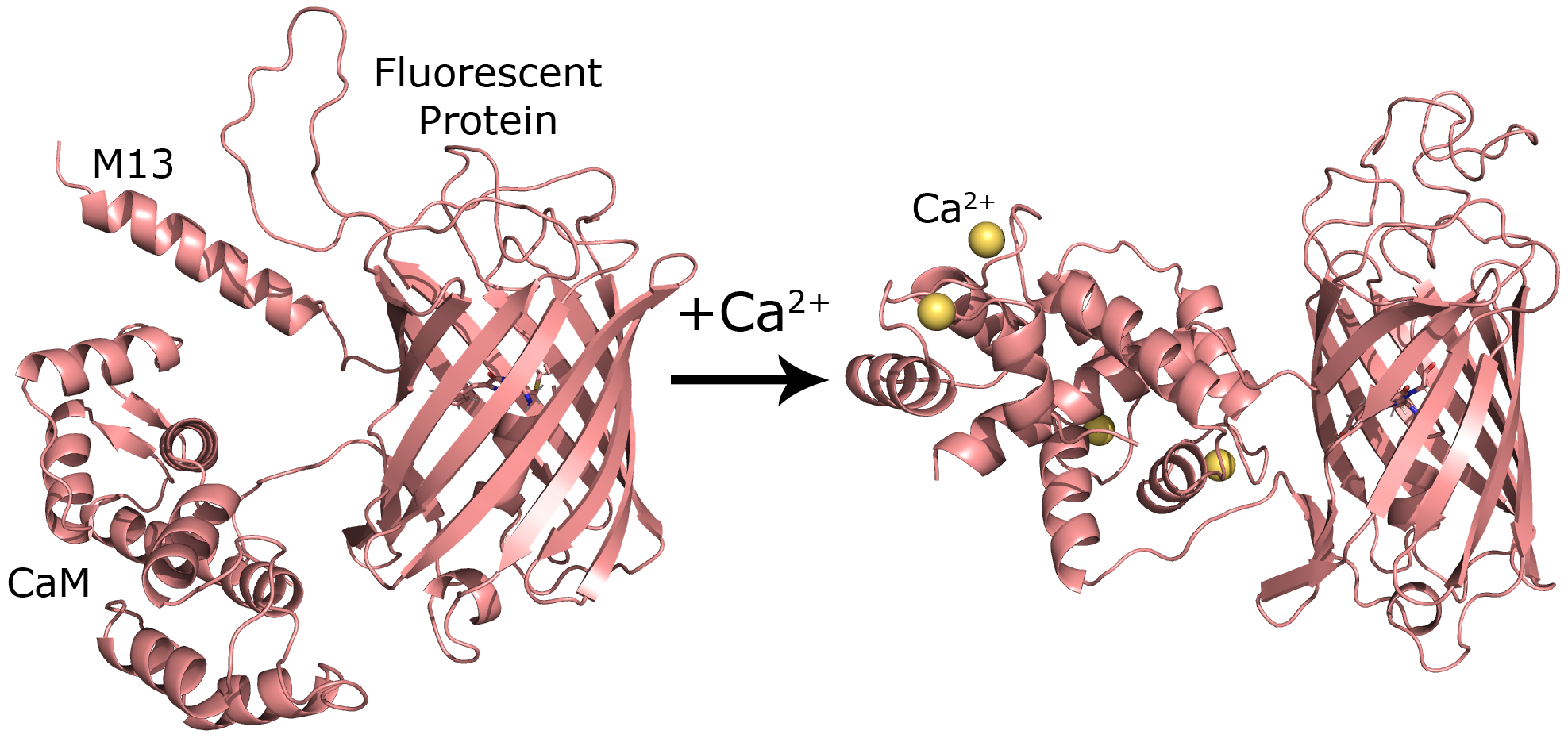
When calcium ions bind to CaM, its structure changes drastically, and this change also impacts the adjacent fluorescent protein region, causing its photochemistry to change. To determine what these structural differences are, I created 5 structural predictions for the protein with and without calcium. REX-GECO does not have a structure that has been determined from experiments. Instead, I relied on homology modeling with the MODELLER software to generate structural predictions. Homology modeling is a method for generating predictions for unknown protein structures using structures from known structures. This software takes one or more protein structures along with their amino acid sequences and uses this information to predict the structure of a target protein.
Molecular Dynamics Simulations
Next, I performed molecular dynamics (MD) simulations on several of these generated structures. These simulations provide many structural snapshots of the protein, which I then used to predict aggregate structural properties. Each of these simulations was carried out for 25 million steps, and a snapshot of the system is saved every 10 thousand steps. Once they completed, I collected the saved snapshots and analyzed them in Python using the MDAnalysis package.
Results
The dihedral angles on the chromophore are important structural features that influence the photochemistry of the protein. The chromophore is the molecular fragment that is responsible for the protein’s fluorescence, and a model of it is shown below.
I measured the distribution of the \(\alpha\) and \(\beta\) dihedral angles shown in the figure, which measure the rotation of the chromophore. After aggregating the measurements across all 10 simulations, I plotted the density estimate of the angle distribution.
On average, \(\alpha\) is −5° when calcium is bound to the protein and 8° when it is not. Similarly, \(\beta\) is 1° and 3° for the same comparison. These distributions show that both the \(\alpha\) and \(\beta\) angles tend to be larger in magnitude when calcium is not bound to the protein. This suggests that the chromophore is more twisted when there is no calcium present. Combining this observation with experimental results, a hypothesis for why the two protein forms differ in their photochemistry emerged: the more twisted state is partly responsible for the large color change between the two protein forms.
My senior thesis provides a more detailed exposition of the project, although the focus and analysis of the project has evolved since it was published.

